Submitted by A.S. Quenault on Mon, 14/04/2025 - 12:03
New research published in Cell Genomics, led by Jennifer Asimit and colleagues, is providing powerful insights into the genetic basis of complex health traits.
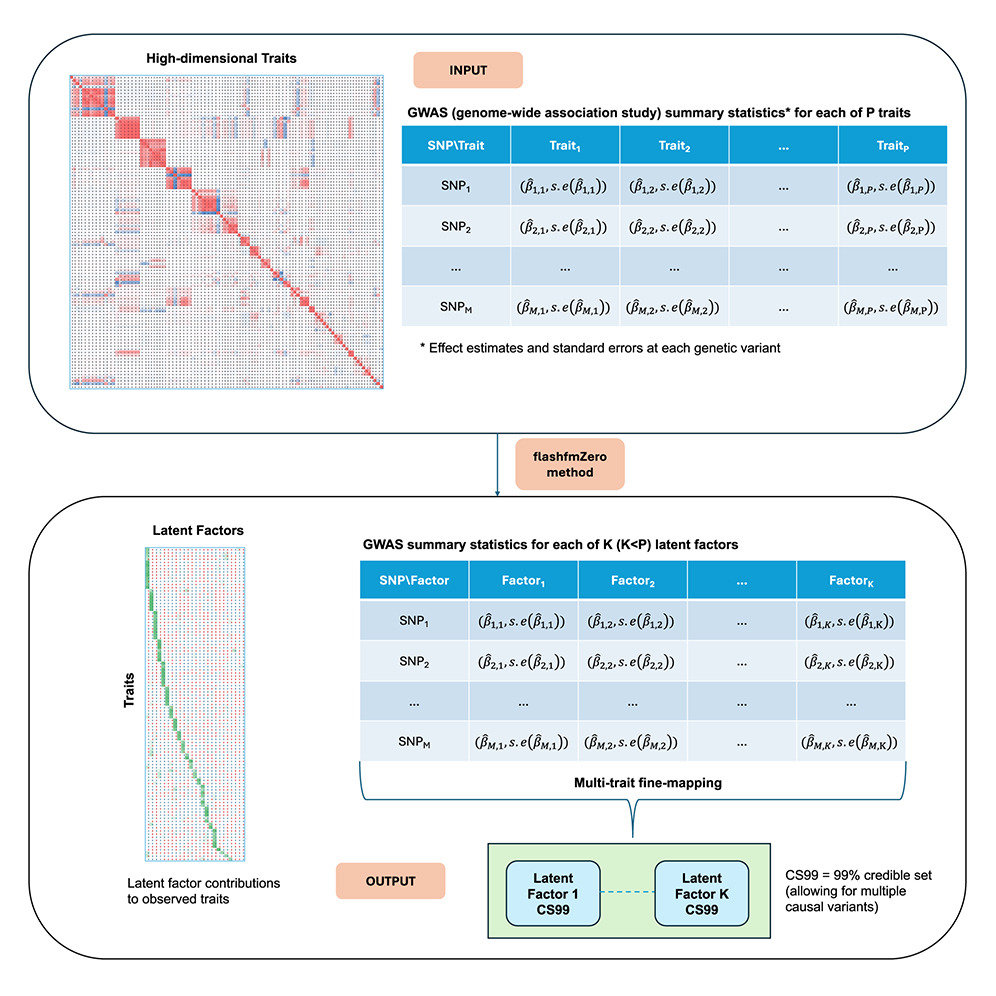
Often a genetic variant affects a group of health-related traits through a common pathway. Methods that leverage information between traits can identify associated variants and pinpoint causal variants more powerfully than methods that analyse traits individually. Such approaches provide an efficient way to gain statistical power without increasing sample size. Partly because most multi-trait methods are limited to a handful of traits, genetic analyses of high-dimensional traits, such as gene expression, protein or metabolite levels, are often analysed one-by-one, ignoring relationships between biologically related traits.
Latent factors cluster traits that have common underlying biological mechanisms, which was the motivation for developing flashfmZero, for analysing many traits together by considering relationships between their underlying latent factors. The new software provides (i) a way to calculate GWAS summary statistics of latent factors from observed trait GWAS summary statistics and (ii) a multi-trait fine-mapping method for jointly analysis of latent factors to pinpoint causal variants that are shared or distinct amongst the factors.
These latent factor approaches were developed and applied to summary statistics of 99 blood cell traits and 184 metabolic traits, by researchers at the MRC Biostatistics Unit (Feng Zhou, William Astle, Jennifer Asimit) and the British Heart Foundation Cardiovascular Epidemiology Unit (Adam Butterworth). All traits were measured in the INTERVAL cohort of UK blood donors. They illustrated that genome-wide significant genetic associations can be identified between latent factors and variants that exhibit only sub-threshold associations with the measured traits. Moreover, flashfmZero tended to produce shortlists of likely causal variants (credible sets) that were equal or smaller than those based on individuals analyses of observed traits.
flashfmZero offers insights into the genetic basis of complex health traits. As it can be applied to GWAS summary statistics and does not require all traits to be measured in all participants, it offers a flexible and efficient approach that boosts power in the analysis of high-dimensional traits. flashfmZero is available as an R library at: https://jennasimit.github.io/flashfmZero/
Dr Jennifer Asimit, Senior Research Associate at the BSU, said:
“flashfmZero provides a powerful and efficient approach to reusing GWAS summary statistics from many traits into a smaller number of biologically meaningful latent factors. These factors have favourable statistical properties that make large-scale joint analyses computationally efficient and more powerful without increasing sample size.”
Read full paper published in Cell Genomics: Improved genetic discovery and fine-mapping resolution through multivariate latent factor analysis of high-dimensional traits - ScienceDirect
This work is funded by the Medical Research Countil, Isaac Newton Trust, Medical Research Foundation, British Heart Foundation, NIHR Cambridge Biomedical Research Centre, BHF Chair Award, Health Data Research UK, and NHS Blood and Transplant.