Submitted by A.S. Quenault on Thu, 01/08/2024 - 11:50
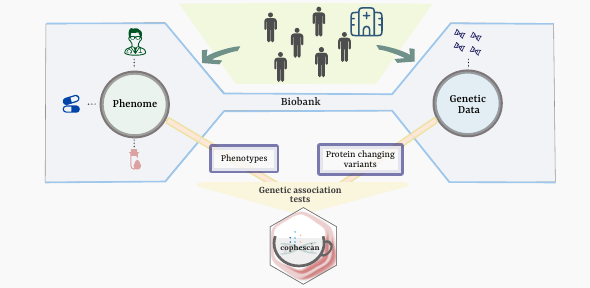
The availability of genetic data linked to electronic health records in large-scale biomedical databases or biobanks has facilitated the widespread popularity of Phenome-Wide Association Studies (PheWAS). These studies explore links between changes in DNA sequence and the “phenome”: patient's characteristics such as disease, laboratory measurements captured through electronic health records and imaging data.
There is now considerable but incomplete knowledge about how genetic variation can alter the expression of specific proteins. A genetic variant associated with a change in protein level eg., in blood, can potentially be linked to disease risk to understand what diseases that protein might be involved in. However, a phenomenon called “linkage disequilibrium” means that genetic variants tend to be correlated with their neighbours, so the variant associated with the protein level may appear to be associated with a disease as a proxy of a neighbouring variant that actually is causally associated with the disease. Thus, a large number of disease-variant links identified by conventional PheWAS methods turn out to be false-positive cases and a separate analysis is required to confirm these findings.
This motivated the development of Coloc adapted Phenome-wide Scan, (CoPheScan), a one-step method for PheWAS that addresses previous limitations, recently published in Nature Communications. It probes causal links between variants and diseases and surpasses the capability of conventional PheWAS tests by distinguishing whether a variant is causal or merely correlated with the actual causal variant.
CoPheScan was developed by Ichcha Manipur from the University of Cambridge and Chris Wallace from the MRC Biostatistics Unit, and scientists from GSK and MSD, as part of an industry-academic collaboration. As CoPheScan identifies associations between the same variant and multiple phenotypes, it can help uncover underlying biological mechanisms that are common among these phenotypes. This has great potential to facilitate the drug discovery process by aiding in the identification of drug targets and their side effects.
In simulation studies, CoPheScan identified substantially lower false-positive results compared to traditional PheWAS approaches. The researchers then used CoPheScan to test links between variants associated with blood protein levels and and 2275 disease-related phenotypes from the UK Biobank, a large biobank with genetic and phenotype data from 500,000 participants, to demonstrate effectiveness on real-world data. The study identified several known and new links between proteins and disease, including a potential role for the protein TGM3 in skin cancer.
“With this tool, we will be able to better understand which proteins change risk of disease, to identify possible targets for therapeutic intervention. This will only be possible thanks to the work to build biobanks in several different countries across the world, which means we will be able to amass more than a million samples to answer these questions." - Dr Chris Wallace, Senior Author
“We successfully tackled a well-known problem of confounding in PheWAS due to linkage disequilibrium. Using the UK Biobank data, we demonstrated that CoPheScan is scalable to biobank-level analyses, and we are excited to extend the method to explore multiple ancestries across different biobanks." - Dr Ichcha Manipur, First Author
This work is funded by the Wellcome Trust (WT220788), the MRC (MC_UU_00002/4), G.S.K. and M.S.D.

